New Generation of High-Throughput Glycomics
The role of Zoldoš Group (Department of Biology, Faculty of Science) as the project partner:
Laboratory for epigenetics will be involved in this project by production of immunoglobulin G (IgG) in HEK293FreeStyle transient system. We will manipulate candidate genes, previously associated with IgG glycosylation by GWAS, using CRISPR/Cas9 tools. This will enable a source of natural and modified glycoproteins needed for development of analytical methods.
Project of Natioanal Centre of Research Excellence in Personalized Healthcare
Laboratory for Epigenetics – Partner in the project
The role of Zoldoš Group (Department of Biology, Faculty of Science) as the project partner:
The project of Personalized Healthcare will provide a platform for internationally recognised institutions to reunion as partners in scientific research, development and innovation in the field of personilised medicine. Laboratory for Epigenetics in Department of Molecular Biology (Faculty of Science) will be involved in the project with investigations of epigenetic regulation of IgG glycosylation. Alternative glycosylation of immunoglobulin G (IgG) can convert this molecule from anti-inflammatory to pro-inflammatory antibody. In a such a way, IgG is involved in inflammatory diseases such as inflammatory bowel diseases, systemic lupus or rheumatoid arthritis. We will use CRISPR/Cas9 tools for targeted gene knock-outs in hybridoma cell line (a model for B lymphocytes). Consequently, our partners in Genos will analyse IgG glycosylation phenotype. The aim is to understand how glycosylation of IgG is established in early B cell life.
Comprehensive Toolbox for Epigenetic Modulation of Gene Expression
In this project, we propose creating a comprehensive molecular toolbox harnessing excellent targeting properties of dCas9 and using it to deliver domains for activation (VPR), repression (KRAB), DNA methylation/demethylation (DNMT3a and TET1) and histone modifications (p300-HAT for histone acetylation and G9a for histone methylation) to promoters or other control elements of targeted genes. The molecular toolbox will be highly modular and easily reconfigured for a range of applications in gene regulation and epigenome editing. We will also functionally validate the assemblies based on Cas9 orthologs and different functional domains, together with selection markers (either for antibiotic selection or fluorescent proteins). A reasonable list of combinations will be tested and this is enabled by modularity of our system. Validation of the epi-CRISPR/dCas9 tools will be done by targeting our candidate loci, relevant for IgG glycosylation, in HEK293 cells and HEK293 Free Style transient expression system, designed to produce high quantity of IgG. The objective is to understand fundamental epigenetic questions such as stability and propagation of the introduced epigenetic marks on the candidate loci.
Type of funding scheme: CRP-Collaborative Research Programme
Proposal number: CRP/HRV17-03
Duration: 24 months (1stst January 2018 – 31st December 2019)
Host institution: Faculty of Science University of Zagreb
Leader of the project: Prof Vlatka Zoldoš, PhD
EU Funding to Faculty of Science: 26,000.00 Eur
Detailed information: https://www.icgeb.org/research-grants.html
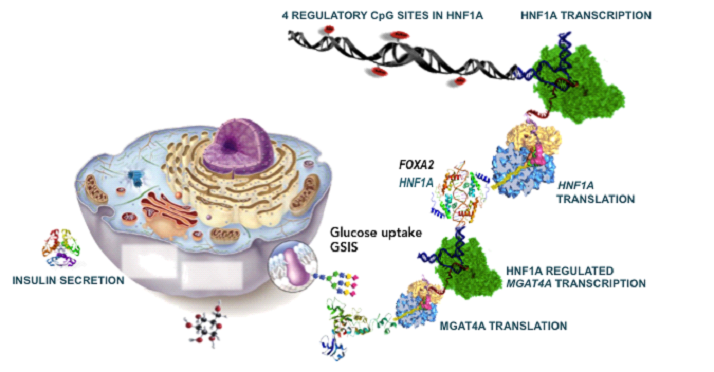
Epigenetic deregulation of the HNF1A gene in diabetes and pancreatic adenocarcinoma
(funded partially through HRZZ project EpiGlycoIgG)
Molecular mechanisms leading to diabetes are not yet fully understood. It is know from studies on mice that high fat diet decreases expression of the HNF1A gene, which is normally highly expressed in liver and pancreas. The transcription factor HNF1A is involved in glucose uptake and proper GSIS (Glucose Stimulated Insulin Secretion) through regulation of the glyco-gene MGAT4A. This gene encodes for glycosyltransferase GntIV which is responsible for correct glycosylation of the Glut transporter on β cells and its localization at cell surface, where it mediates glucose uptake. Similar mechanism is supposed to occur in human Langerhans islets. Our previous results have revealed the correlation between DNA methylation from blood and plasma glycans in patients with HNF1A-MODY patients (a subtype of type 2 diabetes caused by HNF1A mutation) (Zoldoš et al. 2012 Epigenetics).
It has been postulated that a common comorbidity of diabetes is pancreatic adenocarcinoma. The aim of our research is to analyze CpG methylation in the HNF1A gene promoter in β cells of Langerhans islets in pancreas of people who had adenocarcinoma and were previously diagnosed with diabetes. Methylation analysis will be performed using pyrosequencing after bisulfite conversion of DNA extracted from microdissected pancreatic tissue embedded in paraffin. Total glycans from β cells of Langerhans islets of normala tissue as well as in tumor tissue will be determined using Nano LC-MS in order to establish whether HNF1A gene regulates N-glycosylation in diabetes and pancreatic cancer, respectivelly. In addition, putative regulatory sites in the HNF1A promoter will be modulated using CRISPR/Cas9-DNMT3A tool in HCC tumor cell line, the HepG2. N-glycans from secreted glycoproteins will be analyzed using HILIC and MS by our partners.
Completed projects:
Integrated approach to social and economic reintegration of drug addicts
(The project is funded through IPA-CLOUD project; collaboration with prof. Darko Roviš, and prof. dr. sc. Tihana Lenac Roviš, Center for proteomics, Faculty of Medicine University of Rijeka, Croatia)
Genetic and epigenetic variability in the serotonin transporter gene, the SLC6A4, has been previously linked with vulnerability to stress-related psychopathology upon exposure to environmental adversity. This project deals with genotyping the adolescents with risk behaviour (drug addiction) for the SLC6A4 gene represented in the human population with three alleles – S, LG and LA – of which alleles S and LG are associated with predisposition to drug addiction. Our group aims to study DNA methylation profile within the SLC6A4 gene in the cohort of young addicted individuals compared with healthy individuals.